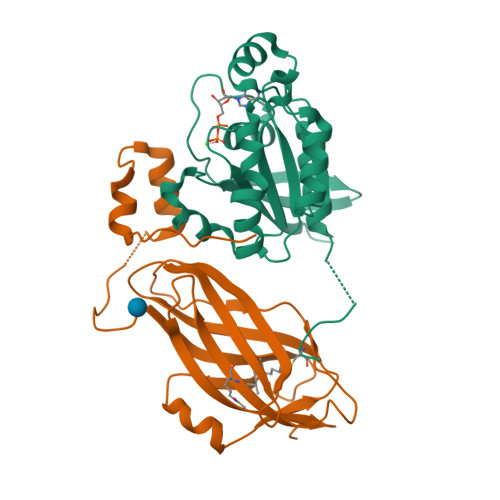
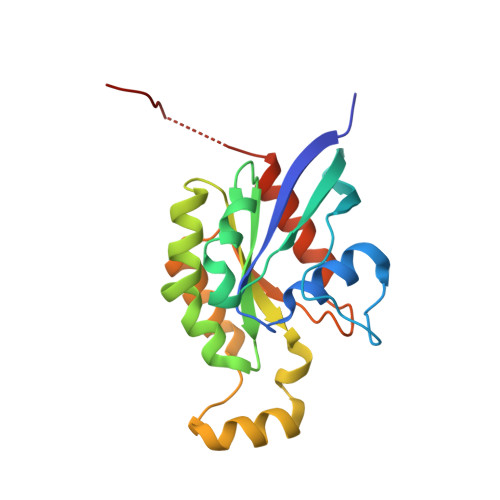
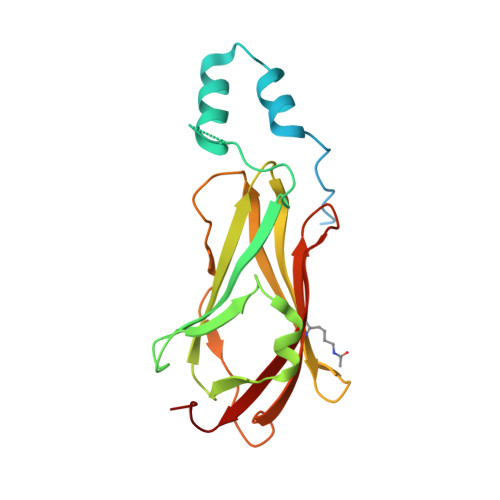
Structural and Mechanistic Insights Into the Regulation of the Fundamental Rho-Regulator Rhogdi Alpha by Lysine Acetylation.
Kuhlmann, N., Wroblowski, S., Knyphausen, P., De Boor, S., Brenig, J., Zienert, A.Y., Meyer-Teschendorf, K., Praefcke, G.J.K., Nolte, H., Kruger, M., Schacherl, M., Baumann, U., James, L.C., Chin, J.W., Lammers, M.(2016) J Biological Chem 291: 5484
- PubMed: 26719334
- DOI: https://doi.org/10.1074/jbc.M115.707091
- Primary Citation of Related Structures:
5FR2 - PubMed Abstract:
Rho proteins are small GTP/GDP-binding proteins primarily involved in cytoskeleton regulation. Their GTP/GDP cycle is often tightly connected to a membrane/cytosol cycle regulated by the Rho guanine nucleotide dissociation inhibitor α (RhoGDIα). RhoGDIα has been regarded as a housekeeping regulator essential to control homeostasis of Rho proteins. Recent proteomic screens showed that RhoGDIα is extensively lysine-acetylated. Here, we present the first comprehensive structural and mechanistic study to show how RhoGDIα function is regulated by lysine acetylation. We discover that lysine acetylation impairs Rho protein binding and increases guanine nucleotide exchange factor-catalyzed nucleotide exchange on RhoA, these two functions being prerequisites to constitute a bona fide GDI displacement factor. RhoGDIα acetylation interferes with Rho signaling, resulting in alteration of cellular filamentous actin. Finally, we discover that RhoGDIα is endogenously acetylated in mammalian cells, and we identify CBP, p300, and pCAF as RhoGDIα-acetyltransferases and Sirt2 and HDAC6 as specific deacetylases, showing the biological significance of this post-translational modification.
Organizational Affiliation:
From the Institute for Genetics and Cologne Excellence Cluster on Cellular Stress Responses in Aging-associated Diseases (CECAD), Joseph-Stelzmann-Strasse 26, University of Cologne, 50931 Cologne, Germany.